Every day the global healthcare system generates tons of medical data that — at least, theoretically — could be used for machine learning purposes. Regardless of industry, data is considered a valuable resource that helps companies outperform their rivals, and healthcare is not an exception.
In this post, we’ll briefly discuss challenges you face when working with medical data and make an overview of publicly available healthcare datasets, along with practical tasks they help complete.
Medical Data: What to Consider When Working with Healthcare Information
The main problem with healthcare data is its sensitivity. It contains confidential information which is protected by the Health Insurance Portability and Accountability Act (HIPAA) and cannot be used without special consent. In the medical sphere, sensitive details are called protected health information or PHI.
Protected Health Information and HIPAA identifiers
Protected Health Information (PHI) resides in various medical documents like emails, clinical notes, test results, or CT scans. While diagnoses or medical prescriptions per se are not considered sensitive, they become subject to HIPAA when linked to so-called identifiers — names, dates, contacts, social security and account numbers, full-face photos, and other elements by which you can locate, contact or identify a particular patient.
There are 18 HIPAA identifiers, and even one is enough to make an entire document PHI that can’t be disclosed to third parties. Read our article on HIPAA violations to avoid common mistakes and associated penalties.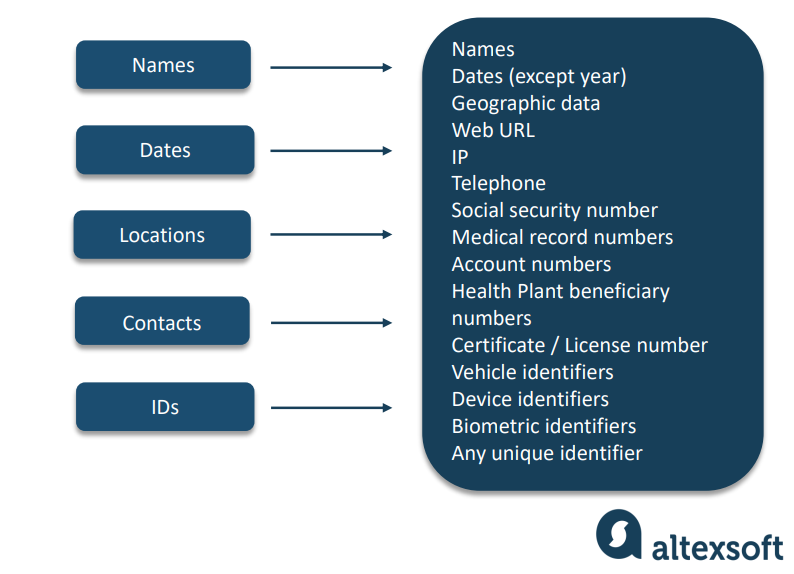
All categories of PHI Identifiers with examples
Medical data anonymization and de-identification
So, before you can use medical data for research or business purposes, you must get rid of personal identifiers and even parts of them (like initials). There are two ways to do it — anonymization and de-identification.
Anonymization means removing all sensitive data forever. At the same time, de-identification only encrypts personal details and hides them in separate datasets. Later, identifiers can be reconnected with health information. The second method takes more effort than the first. Yet, in both cases, the need to comply with regulations adds an extra step to the preparation of datasets for machine learning.
Medical data labeling
Medical or not, unstructured data — like texts, images, or audio files — require labeling or annotation to train machine learning models. This process involves adding descriptive elements — tags — to pieces of data so that a computer can understand what the image or text is about. Read our article on how to organize data labeling to learn about annotation tools and best practices.
Speaking of healthcare data, the labeling must be done by medical professionals. Their services cost significantly more per hour than that of annotators without domain expertise. This creates another barrier to generating quality medical datasets.
Let’s sum up. Though medical data is abundant, getting it ready for machine learning usually takes more time and money than average across other industries — due to strict regulations and the engagement of highly-paid domain experts. No wonder publicly available health datasets are relatively rare and attract much attention from researchers, data scientists, and companies working on medical AI solutions. Below, we’ll explore data collections the Internet has to offer and the practical tasks they help solve.
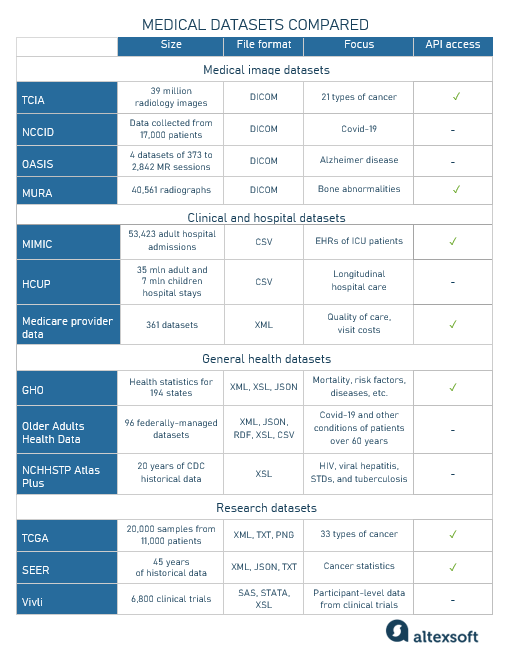 Medical datasets comparison chart
Medical datasets comparison chart
Medical image datasets
Images make up the overwhelming majority (that’s almost 90 percent) of all healthcare data. This provides many opportunities to train computer vision algorithms for healthcare needs. It’s worth noting that medical image data is mostly generated in radiology departments in the form of X-Ray, CT, and MRIs scans. The international healthcare standard for storing and transmitting diagnostic imaging is DICOM (Digital Imaging and Communication in Medicine)
DICOM ensures the imaging equipment produced by different companies speaks the same language and can freely exchange data. Usually, one DICOM file contains several images, technical details, and a header with the patient name and demographics. It goes without saying that in datasets for machine learning all personal details are removed to comply with HIPAA.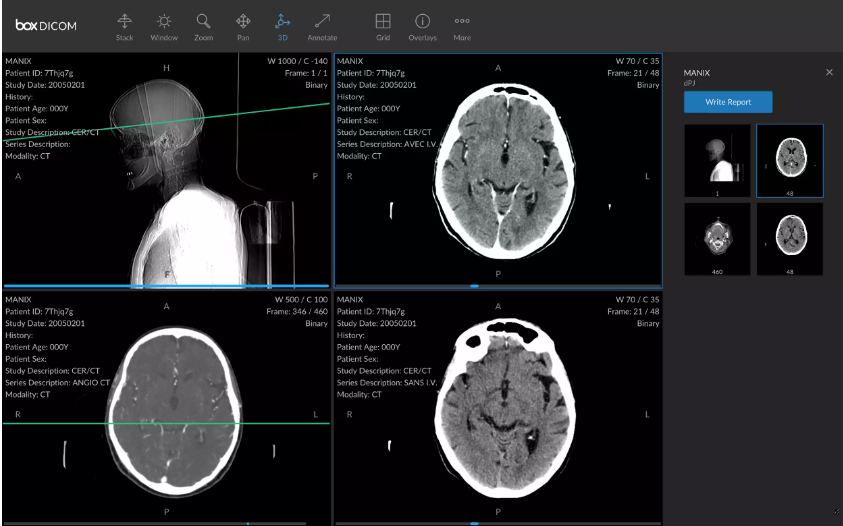
A DICOM file with images and metadata, Source: HIT Consultant
Now, let’s focus on the largest sources of ready-to-use imaging datasets that support AI-powered projects and are of great value to the research community, machine learning developers, and medical professionals.
The Cancer Imaging Archive (TCIA)
Funded by the US National Cancer Institute (NCI), the Cancer Imaging Archive (TCIA) is an open-access hub of de-identified radiology and histopathology images, mostly in DICOM format, representing 21 types of cancer. The data collections are divided by disease (breast cancer etc.), image modality (CT, MRI, and more), or focus of research The archive also augments images with available supporting data like treatment details, patient outcomes, and genomics.
There are four ways of accessing TCIA data:
- by browsing a list of all collections;
- through the Radiology and Histopathology Data Portals with advanced searching and filtering functionality; and
- via TCIA programmatic interfaces (REST APIs) developers can integrate with their apps to establish direct access to data.
The imaging data present in the archive is widely used in scientific research. For example, a recent study aims at assessing the accuracy of deep learning algorithms in the diagnosis of human papillomavirus (HPV) in CT images of advanced oropharyngeal cancer (OPC).
The extended list of scientific publications using the TCIA archive can be found on the official website.
Sample of MRI scans from the TCIA.Source: Cancer Imaging Archive
National Covid-19 Chest Imaging Database (NCCID)
Part of the National Health Service (NHS) AI Lab, the National Covid-19 Chest Imaging Database (NCCID) contains CXR (chest X-ray), MRI, and CT scans from patients in hospitals across the United Kingdom. It’s one of the largest archives of this type, with 27 hospitals and trusts contributing to it.
The training data collected from more than 17,000 patients can be used for the development of image processing software, mathematical modeling, and validation of AI products. For example, it enables you to build a model that forecasts COVID-19 risk score from chest X-rays or create an AI-fueled decision support tool to help identify coronavirus on a particular scan.
Open Access Series of Imaging Studies (OASIS)
OASIS Brains database aims at making neuroimaging data freely available to the scientific community. This extensive data hub exists thanks to research groups and centers at Washington and Harvard universities and the Biomedical Informatics Research Network (BIRN).
Sample images from the OASIS dataset. Source: Researchgate
OASIS contains four datasets: MRI, CT, and PET (positron emission tomography) scans from patients aged 18 to 96 years. These collections support creating data-driven diagnostic tools for Alzheimer's disease. Yet, to access to images, you have to explain to the OASIS team how you’re going to use them.
Musculoskeletal Radiographs (MURA)
MURA (Musculoskeletal Radiographs) is one of the largest public datasets of X-ray images. It contains 40,561 radiographs of upper extremities (like forearms, elbows, shoulders, etc) from 14,863 studies, involving 12,173 patients. Board-certified radiologists from Stanford Hospital manually labeled each study as normal or abnormal.
All MURA data becomes available for download after registration on the webpage.
Today, when over 1.7 billion people worldwide suffer from severe musculoskeletal conditions, such a collection can facilitate the use of deep learning in medical diagnosis, which, in turn, enables spotting the disease at the earliest stages and helps solve the problem of the radiologist shortage.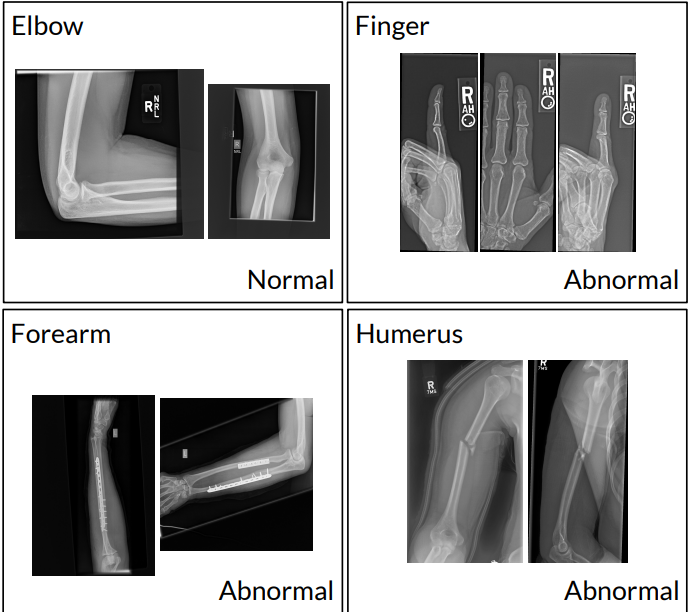
X-rays of upper extremities labeled as normal or abnormal. Source: MURA
Clinic and hospital datasets
Hospitals produce tons of medical and, specifically, patient information, from vital signs and treatment plans to payments and readmissions. The major part of this data resides in the internal systems of healthcare facilities — namely, EHR (Electronic Health Record) systems, medical practice management systems, laboratory information systems, patient portals, and more.
Other sources of care-related information are medical billing tools and databases of government healthcare agencies. Properly structured and organized, these massive collections could drive machine learning projects. Let’s see where to find the largest ready-to-use hospital datasets which highlight different aspects of healthcare.
Medical Information Mart for Intensive Care (MIMIC)
MIMIC is the largest publicly available collection of de-identified electronic health records (EHRs) related to intensive care unit (ICU) patients. Managed by MIT Laboratory for Computational Physiology, it includes information on admission details, prescribed medications, vital signs, laboratory measurements, and more.
MIMIC has attracted strong interest from the ML community that works on solving different medical problems. A recent study based on MIMIC describes the training of a reinforcement learning model to help select treatment for sepsis. Other research focuses on the development of decision support tools to detect ready-for-discharge ICU patients.
Healthcare Cost and Utilization Project (HCUP)
HCUP, run by the Agency for Healthcare Research and Quality (AHRQ) hosts nationwide and state-specific databases that can be used to identify and explore trends in healthcare access, utilization, outcomes, and more.
The project includes the US-largest publicly available collections of inpatient care data. It embraces over 35 million hospital stays of adults and over 7 million hospitalizations of children. This data can be employed to train models for predicting the risk of diseases, length of hospital stay, healthcare costs, and more. It’s also a valuable instrument for researching rare diseases.
Medicare provider data
The Medicare Provider Catalog aggregates official data of the Centers for Medicare and Medicaid Services (CMS). It covers numerous topics, from the quality of care across hospitals, rehabilitation centers, hospice agencies, and other healthcare facilities to visit costs to information on doctors and clinicians. You can view the data on the browser, download a particular dataset in the CSV format, or connect your application to the website via API.
General health datasets
General health datasets are usually managed by governmental bodies and international organizations. These data can be useful for exploring health trends, disease research to understand and prevent epidemics, and other tasks.
Global Health Observatory datasets (GHO)
The Global Health Observatory (GHO) is the World Health Organization’s collection of health statistics for 194 member states. It provides datasets structured around various topics — child health, HIV, tuberculosis, immunization, mental health, and nutrition, to name just a few. You can freely download datasets you’re interested in from the website, choosing from the available formats — CVS and Excel tables, XML, and JSON files. All content is also available via Athena API based on modern, REST architecture.
The GHO's target audience is made up of policymakers and public health professionals in member states and various international organizations. Data from the hub is often cited in academic studies dedicated to global health problems.
Older Adults Health Data Collection
Older Adults Health Data Collection on Data.gov consists of 96 federally-managed datasets. Its key purpose is to bring together information about the health of people over 60 years of age in and out of the Covid-19 pandemic context. Among the organizations that maintain the collection are the US Department of Health & Human Services, Department of Veterans Affairs, Centers for Disease Control and Prevention (CDC), and others. Datasets are available for download in different formats: HTML, CSV, XSL, JSON, XML, and RDF.
NCHHSTP AtlasPlus
NCHHSTP AtlasPlus gives access to 20 years of historical data on human immunodeficiency (HIV), viral hepatitis, sexually transmitted diseases (STD), and tuberculosis (TB). The collection is maintained by the Centers for Disease Control and Prevention: For the most part, they take advantage of data from the American Community Survey. AtlasPLus, in particular, uses the five-year estimates that cover all of the US counties. The information is available in the form of downloadable charts, choropleth maps, and Excel tables.
Research datasets
Research datasets target the scientific community, pharma companies, laboratories, and other entities participating in treatment and drug development. They accumulate information from previous studies to drive medical investigations.
The Cancer Genome Atlas (TCGA)
The Cancer Genome Atlas (TCGA) is a landmark genomics database that covers 33 types of this disease including 10 rare instances. TCGA was founded in 2006 resulting from collaboration between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI).
Since its foundation, the program has produced over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data from 11,000 patients, which is equal to 212,000 DVDs.
TCGA data is available through the Genomic Data Commons Data Portal where you can also find tools for data visualization and analysis. For developers, there is the REST API that gives programmatic access to all data and functionality — including searching, viewing, downloading, and submitting data files, metadata, and annotations.
As for the impact of TCGA, it’s already contributed to the improvement of cancer diagnosis, treatment, and prevention. For example, research based on TCGA enables the identification of tumor subtypes with a distinct set of genomic alterations. Potentially, this will help find the perfect, personalized treatment based on specific genomic changes of a particular patient (Read our article on precision medicine that gives more context to the topic).
The Surveillance, Epidemiology, and End Results (SEER) Program datasets
The Surveillance, Epidemiology, and End Results (SEER) Program is the most reliable source for cancer statistics in the US that aims at reducing the cancer burden within the population. Its database is supported by the Surveillance Research Program (SRP) which is a part of NCI’s Division of Cancer Control and Population Sciences (DCCPS).
SEER data is applicable to multiple cases — for example
- examining the stage at diagnosis by race or ethnicity;
- calculating survival by stage or age at diagnosis, size of the tumor, and
- determining incidence rates and trends for different cancer sites over time.
You can employ SEER Stat Software to analyze SEER and other cancer-related datasets.
Vivli clinical research datasets
Vivli is a non-profit organization that coordinates, facilitates, and promotes sharing and scientific use of clinical research data. At present, Vivli hosts 6,800 clinical trials and enjoys support from 44 members (world-known companies including AstraZeneca, Bayer, Pfizer, Roche, and more). Its resources attract over 3 million participants from 115 countries.
Vivli runs a data-sharing and analytics platform where you can search for information on available studies. Once you find what you need, complete the data request form. After reviewing, you need to sign a data use agreement. Then, you’ll get access to the secure research environment and analytics tools provided by Vivli. Upon request, you can also bring in your own software and algorithms to analyze data. To download a specific dataset you must get approval from the platform.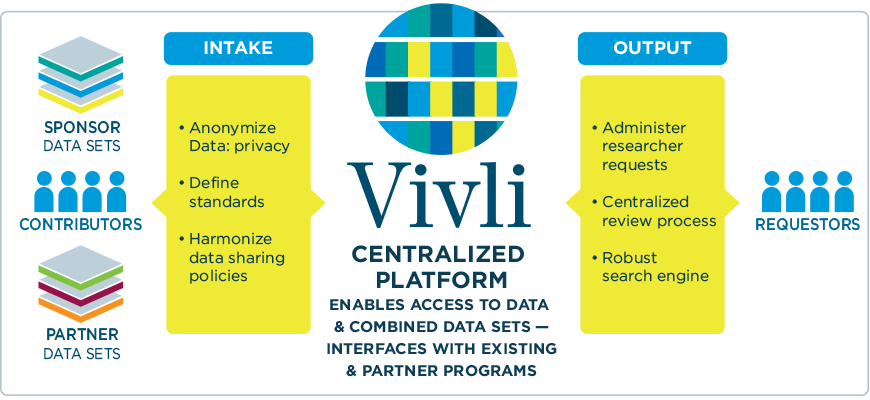
Visualization of Vivli Centralized Platform’s work. Source: MRCT
Where else to look for medical datasets
Besides the specialized repositories we’ve mentioned above, there are other sources of medical datasets for machine learning projects. For example, data.world, a cloud-native data catalog, accumulates almost 3,500 collections related to health. Another platform deserving attention is Papers With Code: It stores 6,964 datasets for ML, with 244 of them relating to the medical domain. You can filter datasets by modality (images or texts), task (classification, segmentation, diagnosis, etc.), and language.
Kaggle which is called an Airbnb for data science also has something to offer. Among its 50,000 public datasets, 953 have tags medical, and over 14, 300 somehow relate to health. AltexSoft used Kaggle datasets of de-identified chest x-rays to build an AI-based lung diagnostics tool that supports decision-making on pneumothorax, pneumonia, and fibrosis. Though it’s always better to have large datasets created for a specific purpose, publicly available resources could still be a perfect fit for small projects and research.

