Subtle at early stages, the signs of lung conditions are easy to overlook. And delays in diagnosis often lead to harsh consequences. That’s where human expertise can be backed by computer vision to create a life-saving collaboration.
In this article, we’ll share key take-aways from our recent experience in building a prototype of a decision support tool that performs three tasks:
- lung segmentation,
- pneumothorax detection and localization, and
- classification of lung diseases.
What is computer vision and how does it benefit health organizations
Computer vision is a subset of artificial intelligence that focuses on processing and understanding visual data. Its ultimate goal is to make machines distinguish objects as living beings do, by simulating the processes of human perception.

Watch this video to make sense of computer vision technology
Mathematical models still can’t rival a person’s sight. But they can act as a second pair of eyes, both tireless and scrupulous. In this capacity, computer vision finds a broad application in healthcare — an industry, where the speed and accuracy of decisions are often a matter of life and death.The list of improvements brought by AI-backed technology embraces
- early disease recognition,
- more accurate image interpretation,
- accessibility of diagnostics in a variety of clinical settings,
- reduced time to diagnosis, and, as a result,
- more effective and cheaper treatment.
Key computer vision applications in healthcare
X-rays, CT scans, MRIs, ultrasound images and other types of medical photographs create fertile soil for development of AI-based tools assisting therapists with detecting different kinds of abnormalities. Let’s briefly explore the most promising use cases.Detection of catheters on radiographs
Catheters and tubes appear to be the second most frequent abnormal findings on radiographs. When wrongly positioned, they can cause severe complications. Unfortunately, in busy clinical settings, significant delays occur between the time of X-raying and the moment when the image ends up in a radiologist’s hand. In this situation, computer vision can be a significant help in detecting misplacements and prioritizing patients with such problems.Different studies show the capability of AI models to identify and localize catheters on radiographs, with the best results achieved for endotracheal tubes and lower detection accuracy for nasogastric and feeding tubes.
Brain tumor segmentation on MRIs
Magnetic resonance imaging or MRI produces the most detailed pictures of soft tissues like the brain and is widely used for diagnosis of brain tumors.The main challenge here is that such tumors can be of different shapes and sizes. The earlier the abnormality is detected, the better chance is for a positive treatment outcome. And machine learning techniques hold the potential of speeding up tumor localization dramatically.
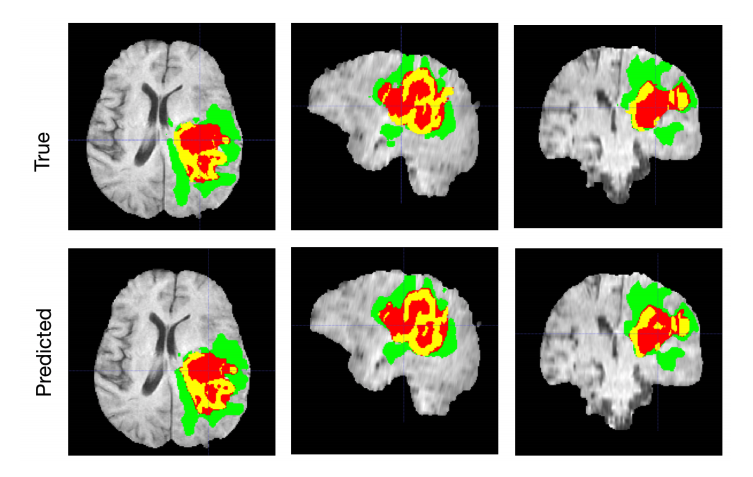
Brain tumor segmentation with AI. Source: Nvidia.Developer
The declared accuracy of various models trained to segment tumors in MRIs ranges from 73-79 to 92 percent. Far from being perfect, AI tools still can be used to support doctors’ decisions.Skin cancer classification on photographs and dermoscopic images
The most widespread oncology disease in the US, skin cancer is more often than not detected visually. Yet, owing to the variability of skin lesions, its symptoms can be confusing. That’s where computer vision outperforms even living experts. Recent research demonstrates that machine learning models show better results in skin cancer classification than an average dermatologist.COVID detection on chest X-rays
Early detection of coronavirus signs from chest X-rays is crucial for preventing the infection from spread. No wonder that extensive research has been made across the globe to test ML models as a diagnostic option. The most advanced AI algorithms achieved an almost 97 percent accuracy.Read our article Deep Learning in Medical Diagnosis to get more information about applications for AI in medical image analysis and barriers to adoption of machine learning in healthcare. Otherwise, let’s proceed to the first and most fundamental step in building AI-fueled computer vision tools — data preparation.
Medical image databases: abundant but hard to access
Computer vision requires plenty of quality data, diverse in gender, race, and geography. But how is a sufficient dataset acquired?While you can prepare a dataset for machine learning from the ground up, this will take resolving the following domain-specific challenges:
- getting ethical approval to access medical image files,
- anonymizing data to comply with regulations, and
- labeling data by medical experts to create a ground-truth dataset.
What is DICOM and why does it need ethical approval
Short for Digital Imaging and Communications in Medicine, DICOM is a global healthcare standard for most types of medical images. It specifies a file format and a communication protocol, ensuring that imaging equipment from different companies speak the same language and can smoothly exchange data. Generally, sticking to DICOM format facilitates achieving interoperability across the entire healthcare industry.A common DICOM file contains
- one to several images,
- a header with patient demographics, and
- technical image details.

A chest X-ray saved in the DICOM format, containing personal information. Source: AWS Machine Learning Blog.
To store and manage DICOM-based content, healthcare organizations use picture archiving and communication systems (PACS) or vendor-neutral archives (VNAs). In both cases, only accredited professionals, such as radiologists or physicians, have access to image repositories.The restrictions come from the fact that DICOM saves protected health information (PHI) like name or age. So, it’s subject to the Health Information Portability and Accountability Act (HIPAA) and the General Data Protection Regulation (GDPR).
How can third-party developers acquire DICOM files? More often than not, the only way is to get "approval from the local ethical committee." Officials access all risks and benefits of a project to grant access to data stored within the hospital environment. After (and if) they give the go-ahead, you can proceed with creating a dataset for computer vision.
Data de-identification / anonymization
Under regulations, all sensitive details that link an image to a particular individual must be removed or hidden before you feed data to your algorithm. De-identification and anonymization are two major approaches required to comply with HIPAA and GDPR.Anonymization means that you just delete (and lose) all PHI. There are many free and commercial tools to automatically wipe off identifying information from the header or replacing it with dummy values. Some examples are DICOM Anonymizer, DICOM Anonymizer Pro, and DICOMAnonymize. See, providers of these tools don’t lose any sleep over creative naming, so you’ll easily find what you need in the Internet!
You can even remove headers without any special software — just convert DICOM files into PNG, JPEG, or TIFF images. These formats are sometimes more convenient as you don’t need a special DICOM viewer to open them on your computer.
De-identification doesn’t erase all the sensitive information forever, but just hides it in separate encrypted datasets. Later, some details can be re-associated with the image. This process is far more complex than anonymization. No wonder that automated de-identifiers are quite rare.
However, even anonymization is not as easy as it may seem. The problem is that images themselves may contain burned-in sensitive data. Optical character recognition (OCR) tools or even human reviews and manual editing are required to get rid of the embedded textual PHI while preserving the image integrity.
It’s worth noting that in research settings, AI developers are sometimes allowed to leave certain PHI elements that can be of use for the project. For this, you must get a HIPAA Waiver of Authorization or patients’ informed consent.
Image labeling by experts
The next big step in data preparation for computer vision is image labeling or annotation. It requires engaging medical experts (in our case, radiologists), which is expensive and time-consuming. Each image should be examined by three professionals to create a quality ground-truth dataset — the one you will use to train and test your model.Another important detail: Image databases must have a clear history of who annotates what to comply with the Food and Drug Administration (FDA) that regulates ML-based solutions in healthcare.
In essence, data labeling involves assigning a special class of metadata to images. This task can’t be executed right in DICOM viewers which often feature basic annotation capabilities. You need professional tagging tools designed for machine learning purposes.
There are two major types of annotations in computer vision:
- semantic segmentation, when radiologists or other experts manually mark and contour an area of interest — for example, an entire organ, a nodule, a tumor, a lesion, etc. These tags enable models to single out an anatomical structure or abnormality from medical images.
- classification when images are categorized into groups, based on the presence or absence of a particular disease. For this task, experts, in most cases, need extra information — like lab test results or prior medical history. Getting access to it and linking it with the image create an additional layer of complexity.

Medical image segmentation of lung nodules performed independently by three experts (their results are marked with different colors). Source: NCBI
Often, two types of annotations are applied simultaneously, so that you can develop a computer vision solution to detect a disease and, if present, segment it. Annotations are stored inside the same DICOM file or as a separate text. Later, it is to be converted to a standard format like JSON or CSV for processing and model development.Finding a public dataset
Chest X-rays and CT scans are the most common imaging procedures, and their quantity per se is not a problem. However, rigorous regulations and high cost of manual annotation create high barriers to obtaining medical datasets. Apparently, their preparation from the ground up is an endeavor for big enterprises and large-scale scientific projects.Smaller AI developers and researchers can save time and money by using open medical imaging databases. Owing to domain-specific obstacles, they are not as plentiful as public datasets for sentiment analysis and other machine learning tasks. Still, chances are you’ll find the right fit for ML experiments, proofs of concept, and narrow-focused decision support tools.
To build our prototype, we resorted to Kaggle, a platform for data scientists that aggregated over 50,000 public datasets. Namely, we used
- Chest Xray Masks and Labels for lung segmentation. The dataset contains de-identified chest x-rays in DICOM format, accompanied by radiology readings in the form of a text file. The images were collected as a part of a tuberculosis control program;
- SIIM-ACR Pneumothorax Segmentation for identifying pneumothorax (air in the pleural cavity) in chest x-rays; and
- NIH Chest X-rays for lung disease classification. The collection contains over 112,000 radiographs from nearly 30,000 unique patients. When assigning disease labels, annotators used Natural Language Processing (NLP) to extract data on abnormalities from the associated radiological reports.
Computer vision algorithms: nets to catch features
Modern computers are capable of categorizing and defining objects due to the subset of machine learning called deep learning. In computer vision, data scientists mostly apply a particular class of deep learning algorithms — convolutional neural networks (CNNs.)Below, we’ll explain how this technology works and what makes it preferable for image recognition tasks.
CNN layers in brief
The multi-layer architecture of CNNs was designed to identify visual features in pixel images with minimum preprocessing. An extremely deep network may contain over 100 layers, but still they will fall into three groups, pivotal for any CNN.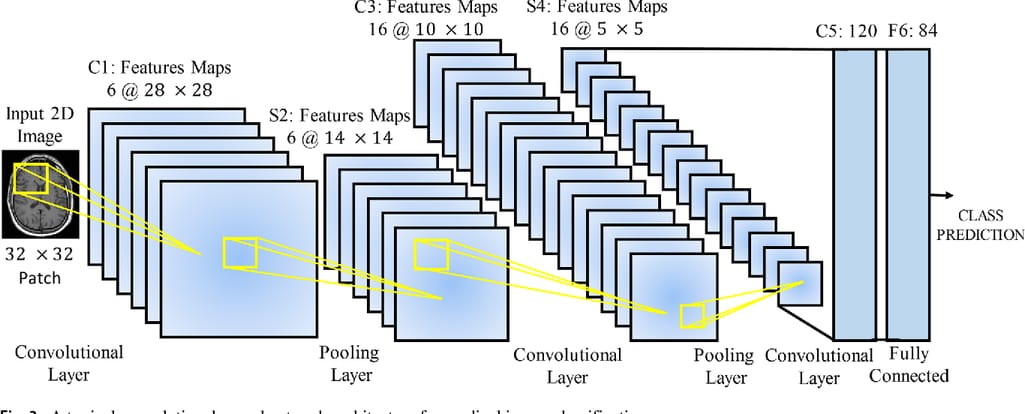
A typical CNN architecture. Source: Frontiers in Neuroscience
Convolutional (conv) layers represent the core part of the CNN. Here, numerous filters known as convolutional kernels slide over an input image, pixel by pixel, to identify different patterns. Typically, early conv layers extract the basic shapes — say, lung edges. The next layers recognize more complex concepts — like nodes or other abnormal structures. The kernels produce multiple output images called feature maps.Pooling layers are inserted in-between and after convolutional layers to compress feature maps, and, consequently, cut the computational power required to process the image.
Fully connected (FC) layers aggregate data produced by previous layers to generate the final output (classification.)
In sum, even extremely complex CNN models can be viewed as a two-component system, made of feature extraction and classification blocks. The conv and pooling layers detect features. FC layers determine the probability of the input image belonging to a certain class.
Choosing the right CNN architecture
After experimenting with different CNNs, we finally went with three architectures that showed the best accuracy as applied to each mentioned-above Kaggle dataset and in performing a certain task.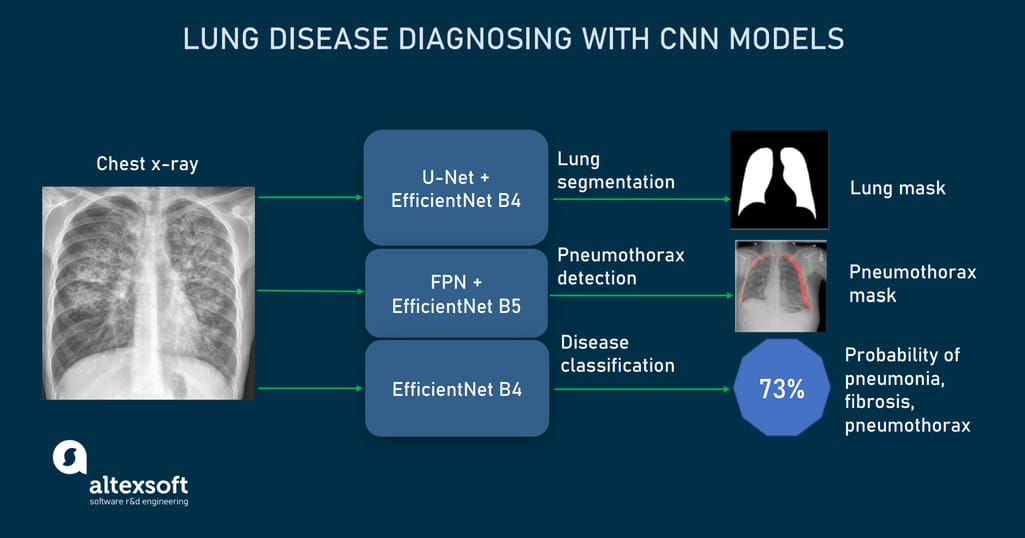
CNN models used to detect and localize lung abnormalities from X-rays.
U-Net for lung segmentation (trained on Chest Xray Masks and Labels dataset). U-net is a CNN architecture designed in 2015 to process biomedical images. It consists of two paths, contractive and expansive, which give the network a U-shape.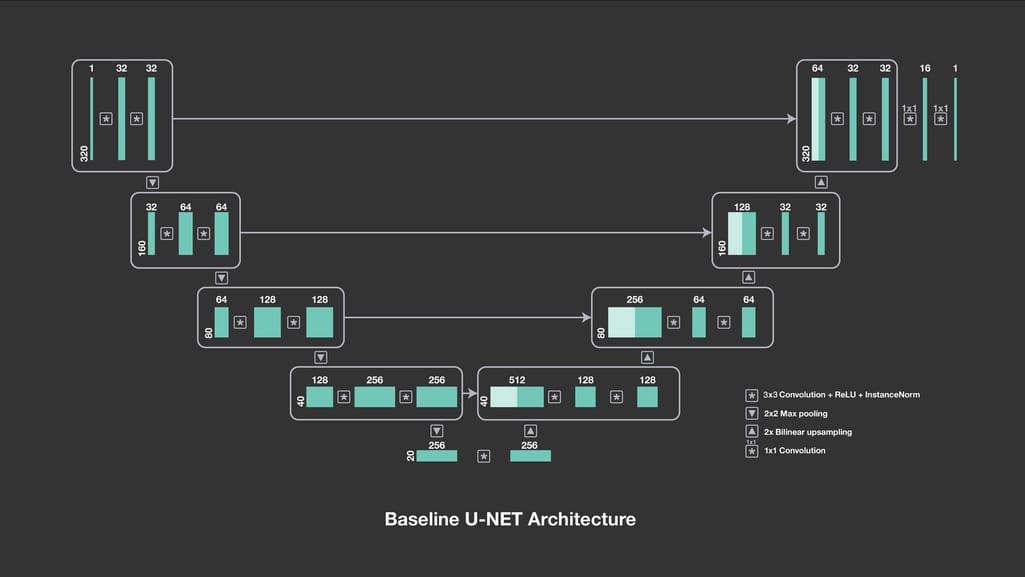
Baseline U-Net architecture. Source: Facebook Engineering
The outputs of contractive layers are copied to corresponding expansive layers, where pooling operations are replaced by their opposites — upsampling operations. Instead of compressing feature maps, upsampling operators increase the number of pixels to preserve the resolution of the input image.This approach allows for more precise segmentation while working with fewer training images. As a result, U-Net not only identifies whether there is a disease but also accurately allocates the problem area. The accuracy we achieved with this network when detecting lung shapes from pulmonary x-rays amounts to 93 percent.
Feature Pyramid Network (FPN) for pneumothorax detection and localization (trained on SIIM-ACR Pneumothorax Segmentation dataset). FPN acts as a feature extractor inside a CNN. Similar to U-Net pathways, it creates two pyramids. The bottom-up pyramid downsamples the input image while top-down one upsamples it.

FPN architecture. Source: Semisupervised Learning for Bone Lesion Instance Segmentation (ResearchGate)
The main difference from U-Net is that FPN not copies outputs of a downsampling pyramid, but passes them through the convolutional layer and makes independent predictions or object detections on each upsampling level (U-Net performs predictions only once, at the end of the expansive path.) The ultimate FPN prediction is computed from all previous predictions. In our case, the FPN-based model had an accuracy of 85 percent.EfficientNet for disease classification (trained on NIH Chest X-Rays dataset). EfficientNet is a comparatively new CNN introduced in 2019 by Google Brain Team. It was designed to maximize classification accuracy without increasing computational cost.
Before EfficientNet, there were three approaches to enhancing the accuracy of a neural network:
- via increasing the model depth — or the number of layers processing the data;
- via increasing the model width —- or the number of filters (kernels) inside each layer; and
- via inputting images of higher resolution, with more fine-grained details.
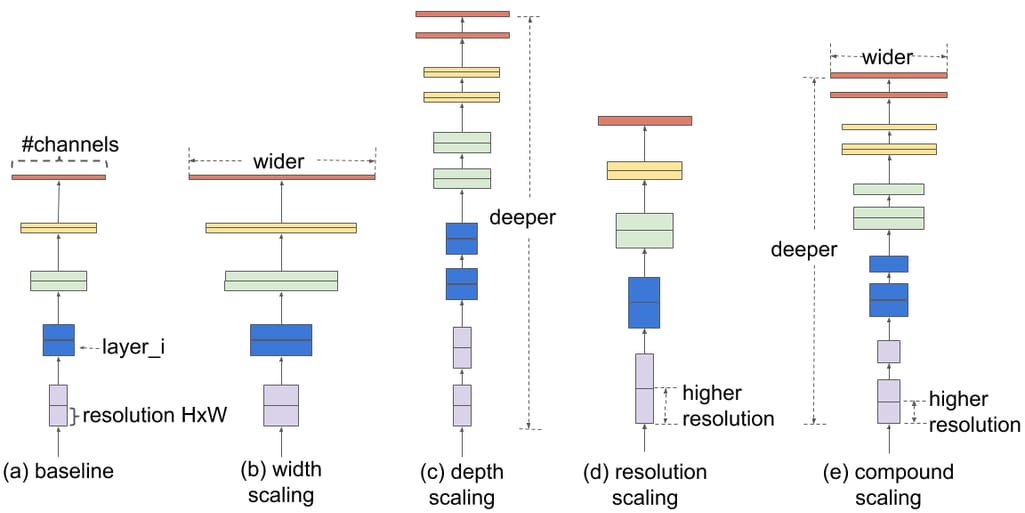
Comparison of different scaling approaches. Source: Google AI Blog
The EfficientNet family consists of eight model versions, from B0 to B7, where each subsequent number reflects more parameters and higher accuracy. For the disease classification part of our project, we decided on the EfficientNet B4 variant.We also used B4 and B5 EfficientNets as backbone networks for the above-mentioned U-Net and FPN models respectively. The backbone defines how layers are arranged in the downscaling block and how the upscaling pathway should be built.
Computer-aided diagnosis solution to detect lung diseases from X-rays
So, our computer vision prototype based on three CNN models allows users — whether a patient or radiologist — to upload a chest X-ray and check the health condition. As output, the tool- creates a lung mask, showing the edges of the organ;
- calculates the probability of pneumonia, fibrosis, and pneumothorax, based on features extracted, and
- if pneumothorax is present, loсalizes it on the lung image.
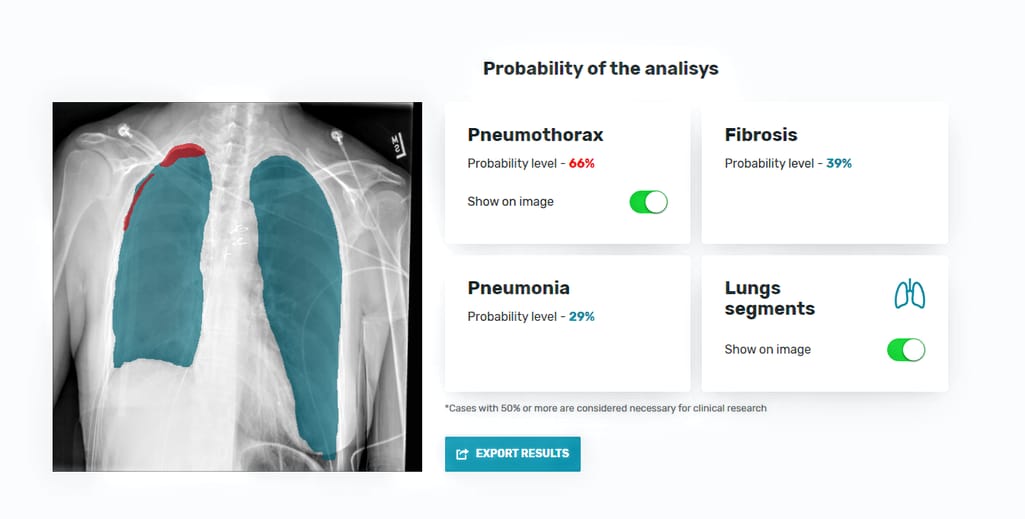
The interface of the AI-based diagnostics tool with results of AI-based X-ray analysis: lung segmentation, pneumothorax localization, and calculation of disease probabilities.
Obviously, the current tool leaves plenty of room for improvement. The possible steps to enhance it are- using larger and more balanced datasets to achieve even better accuracy,
- expanding the pool of lung diseases to be detected, and
- highlighting the area of abnormality on the output images.

Sometimes making predictions from X-rays is just boring. Source: UBC Radiology Twitter
Even with all possible advances implemented, the software by no means is intended to replace a live pulmonologist. It aims at better informing patients and assisting with triaging chest x-rays in clinical settings, reducing reporting delays, and just making the work of diagnostic radiologists somewhat less tedious.