Bioinformatics methods and tools have been growing so fast that soon every biolaboratory might have its own bioinformatician who could derive crucial knowledge about the patient’s molecular system to provide better care. In this article, we want to talk about why omics data analysis is such a popular research topic and different methods of integrating it.
What is omics data and where is it used?
Omics data is information generated by studies ending with -omics: genomics, proteomics, phenomics, etc.It all started with genomics. When the field of genomics first appeared, it was principally different from genetics since it focused on studying the whole genome rather than single genes. Then proteomics, metabolomics, phenomics, and others followed suit, being dedicated to describing the complete protein profile rather than separate proteins or the whole metabolic level instead of one separate metabolite.
So,every area of study whose name ends in -omic belongs to the omics fields of study. This includes the following disciplines:
- genomics (the study of a person’s genome);
- proteomics (the study of proteomes, a set of proteins in an organism);
- metabolomics (the study of metabolites, small molecules within cells produced during metabolism);
- metagenomics (the study of genetic materials recovered from natural environments);
- phenomics (the study of total phenotypic characteristics of an organism);
- transcriptomics (the study of RNA transcripts produced by a genome).
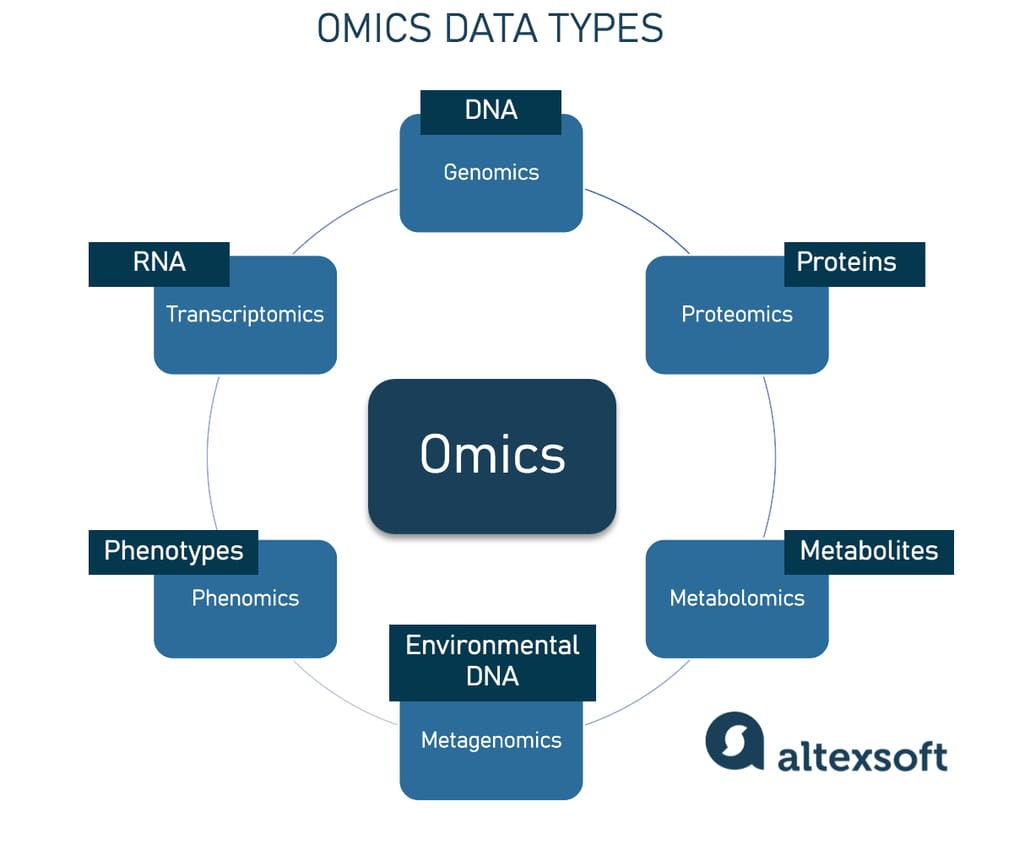
An overview of omics disciplines and what they study
As we already mentioned, data generated by omics studies is called omics data. Compared to a non-omics study, omics research generates a much larger data output. Omics data provides a comprehensive understanding of the underlying structure of a biological system to expand scientific research. For example, omics data generated by the Human Genome Project has been incredibly helpful to the understanding of problems in human disease, agriculture, plant science, microbiology, the environment, and more.The Human Genome Project is an example of a single omics study, where data is analyzed using one assay (method), for example genomics, transcriptomics, or metabolomics. But recently, new tools, technology, and approaches have allowed us to use multi-omics techniques. In multi-omics studies, you can combine datasets of different omics groups for analysis to get a holistic view of human health. This revolutionary shift gives way to truly personalized, precision medicine.
Some of the most prominent examples of omics data use cases involve the following.
Precision medicine
Medicine applies many different methods when providing diagnostics and care, including studying the patient’s medical history, laboratory evaluation, physical examination, and more. One of the ways we can significantly improve the accuracy and efficacy of treatment is by also evaluating the disease on the molecular level.Instead of going through many standard tests, patients with complex cases and rare conditions can get a diagnosis much easier through phenotyping or genome sequencing. At the same time, patient data and samples can be used by researchers in further studies, guiding new discovery and treatment.
Food science and personalized nutrition
The complexity of relationships between human bodies and food makes it very difficult to understand how dietary components would react in an organism. But nutrigenomics – a nascent field of study exploring the relationship between genetics and diet via omics analysis – may help provide truly personalized dietary advice.Not only that, the combination of different omics could become an advanced tool for public health research and human studies that would provide valuable information about the nutritional status of certain populations and develop programs to improve it.
Agricultural biotechnology
Omics technologies have been successfully applied to studying and manipulating nutritive and economic qualities of crop plants. Genome sequencing and genetic mapping have long allowed us to produce better plants such as seedless watermelons and vitamin-packed broccoflowers.Omics technologies enable us to enhance the nutritional properties of food with precision to develop functional products that contain increased vitamin levels and better protein quality to address specific conditions. Since omics can give us insight that we wouldn't have acquired otherwise, we can improve food and energy supply, help preserve the environment, and even solve world hunger in the long run.
Treating age-related diseases and disabilities
Although in the past decades the average life expectancy has dramatically increased, that life hasn’t been disease-free. While achieving older age many people can survive age-related diseases and disabilities, they still suffer from them. Human aging is a complex topic, influenced by diet, gender, socioeconomic status, and, of course, genetics. That’s why we need to look at aging through an integrated lens. Omics data is tasked to finally provide such a view.The employment of multi-omics tools has resulted in the development of methods and platforms for omics data integration, analysis, and visualization. In this article, we will cover some of them that are heavily featured in bioinformatics studies.
But first, let’s review the main sources of multi-omics datasets
Omics dataset repositories
There are a few depositories of publicly available multi-omics patient data that can be used for disease research, diagnostics, and treatment development.The Cancer Genome Atlas (TCGA) – the largest source for cancer omics data
With over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data, the Cancer Genome Atlas is the largest collection of multi-omics datasets spanning 33 cancer types and over 20,000 tumor samples.On the Genomic Data Commons portal that hosts TCGA, you can discover and review over 86,000 conveniently categorized cases, search projects, launch analyses, and, of course, browse and download data.
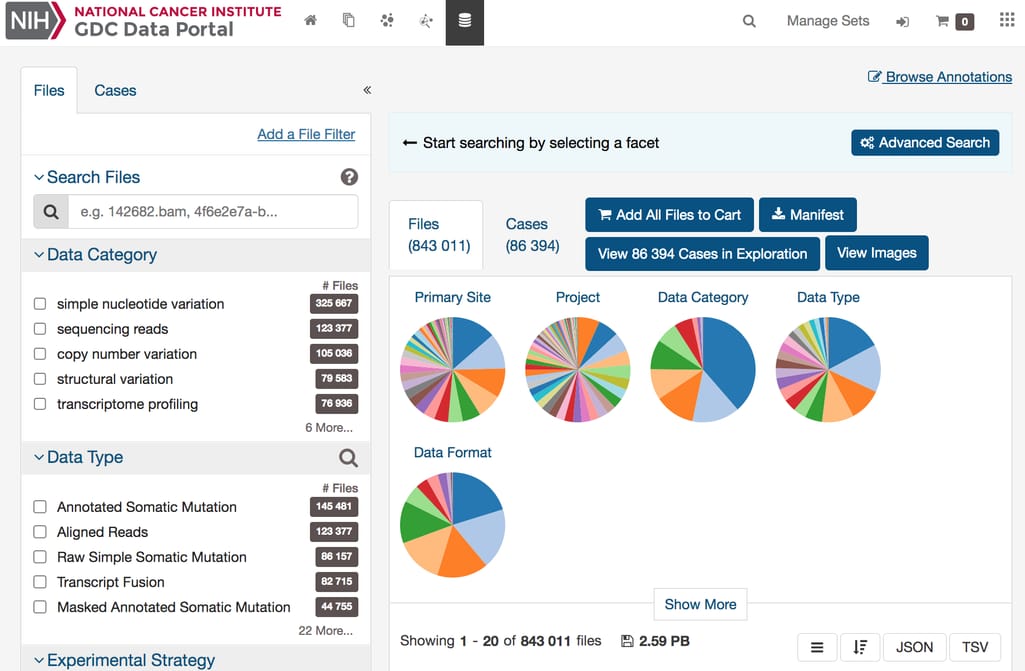
GDC Data Portal interface
TCGA is widely used by the research community and has greatly contributed to discoveries about cancer biology and treatment.TARGET – pediatric cancers assessment source
Another project run by the National Cancer Institute, TARGET aims to provide data for childhood cancer assessment. This data hosts clinical information, gene expression, miRNA expression, and sequencing data of 24 types of cancer.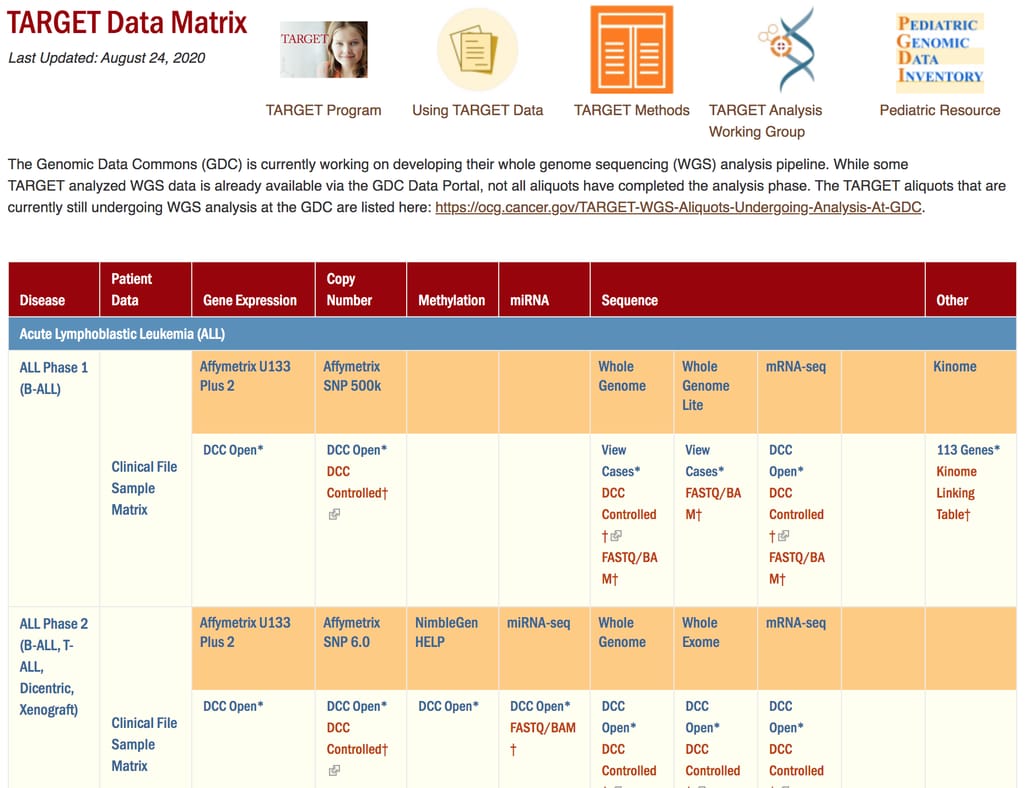
TARGET Data Matrix interface
Some of TARGET data is available on the Genomic Data Commons portal, though many parameters are still undergoing analysis. You can also access it via TARGET Data Matrix.International Cancer Genomics Consortium (ICGC) – cancer mutations datasets
Another source of cancer genomics data is the International Cancer Genomics Consortium. As of February 2021, it doesn’t accept data submissions and currently sits at 86 cancer projects in 22 primary cancer sites from over 22,000 donors. This repository mainly focuses on mutations data.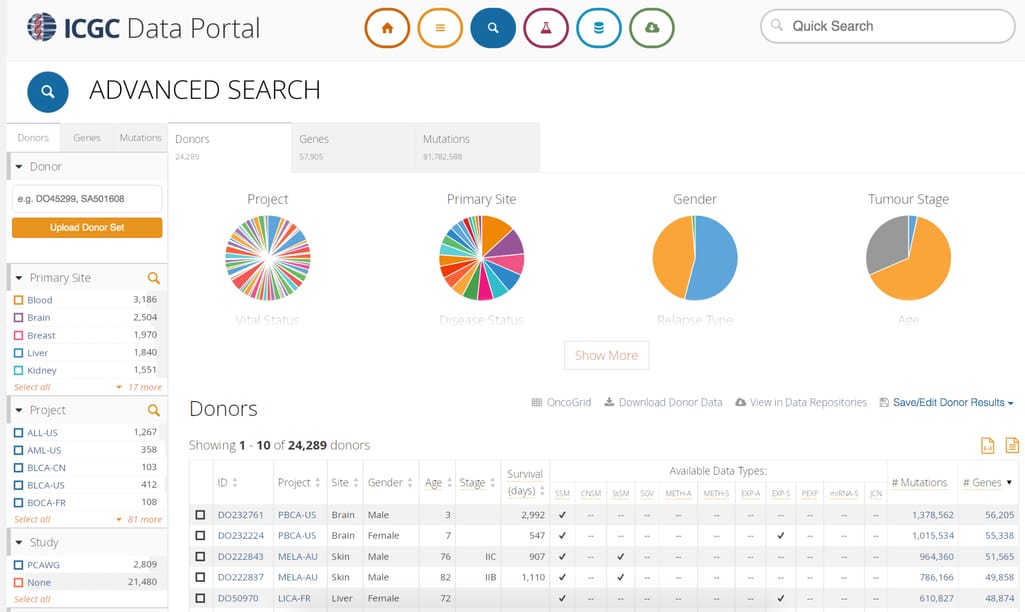
ICGC Data Portal interface
ICGC also hosts data analysis tools, which you can use to run enrichment analysis, cohort comparison, set operations, and visualize genetic alteration on provided data sets or your own gene set.It also has an API to automate the search through ICGC via your own software.
Omics Discovery Index (OmicsDI) - 22 repositories with advanced search
OmicsDI implements a unique search engine for omics datasets with auto complete features and special query syntax that allows users to easily find and filter content. It contains datasets from 22 repositories covering genomics, transcriptomics, proteomics, and metabolomics data.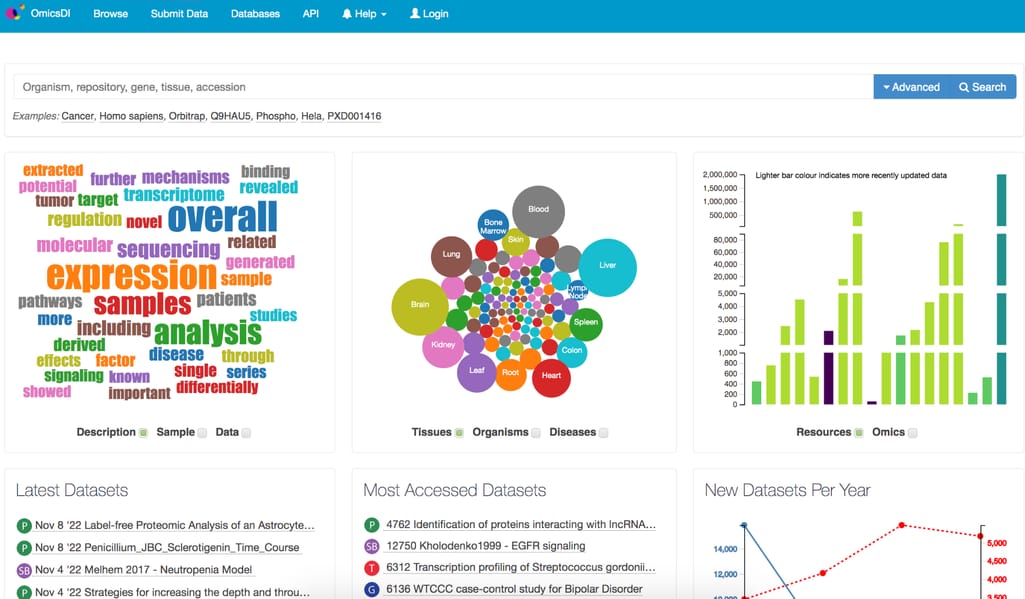
Omics Discovery Index interface
OmicsDI API returns results both in JSON and XML and allows searching, sorting, and filtering data.Finally, let’s talk about how to put this data to use and integrate it for multi-omics analysis.
Multi-omics data integration tools
Today, there are tons of tools and methods available for free and with open access that allow you to integrate omics datasets and derive insights. Namely, you can look into such repositories of tools as Elixir Bio.Tools that has over 500 instruments related to omics analysis on their platform and Bioconductor, which returns over 5500 results on omics data integration packages.Below, we will briefly review some of the most popular ones that you can feel free to explore.
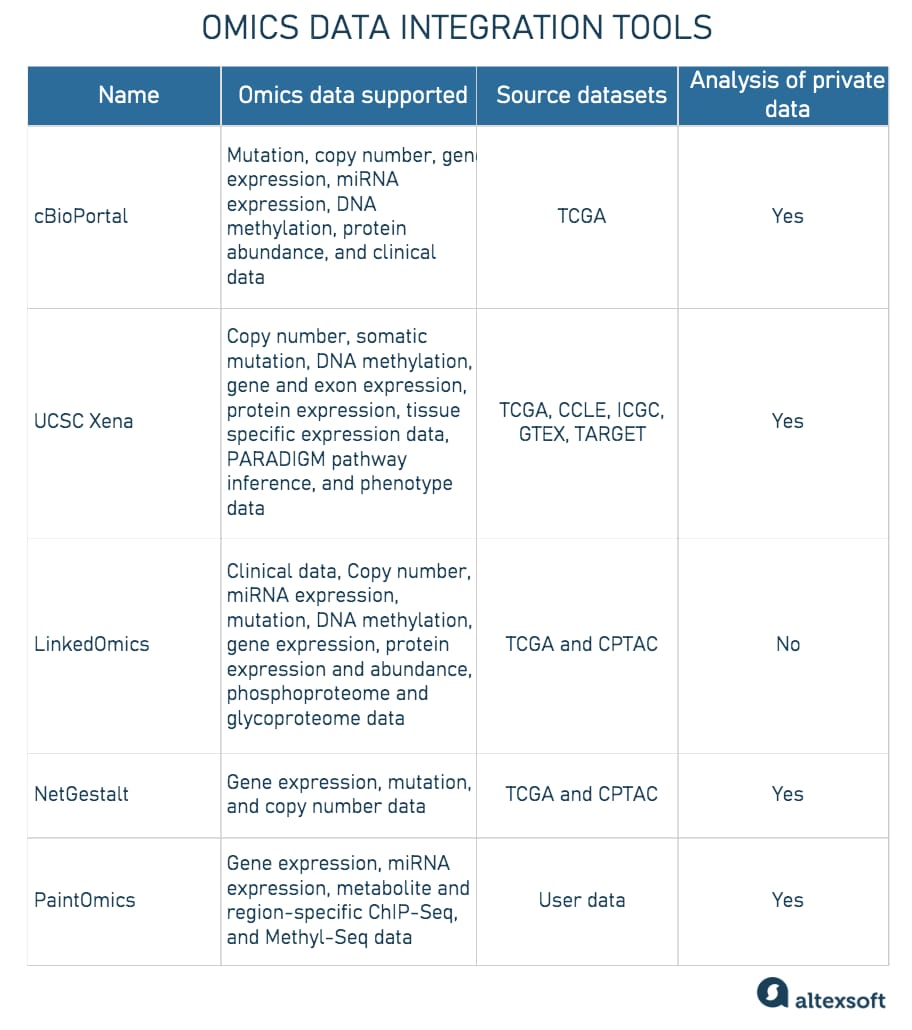 Omics data integration tools
Source: Subramanian I, Verma S, Kumar S, Jere A, Anamika K.
Multi-omics Data Integration, Interpretation, and Its Application
Omics data integration tools
Source: Subramanian I, Verma S, Kumar S, Jere A, Anamika K.
Multi-omics Data Integration, Interpretation, and Its Application
cBioPortal
A user-friendly tool both for bioinformaticians and clinicians, cBioPortal is a platform for exploration of cancer genomics datasets.It currently hosts data from over 200 cancer studies, including:
- non-synonymous mutations,
- DNA copy-number data,
- mRNA and microRNA expression data,
- protein level and phosphoprotein level data,
- DNA methylation data, and
- de-identified clinical data.
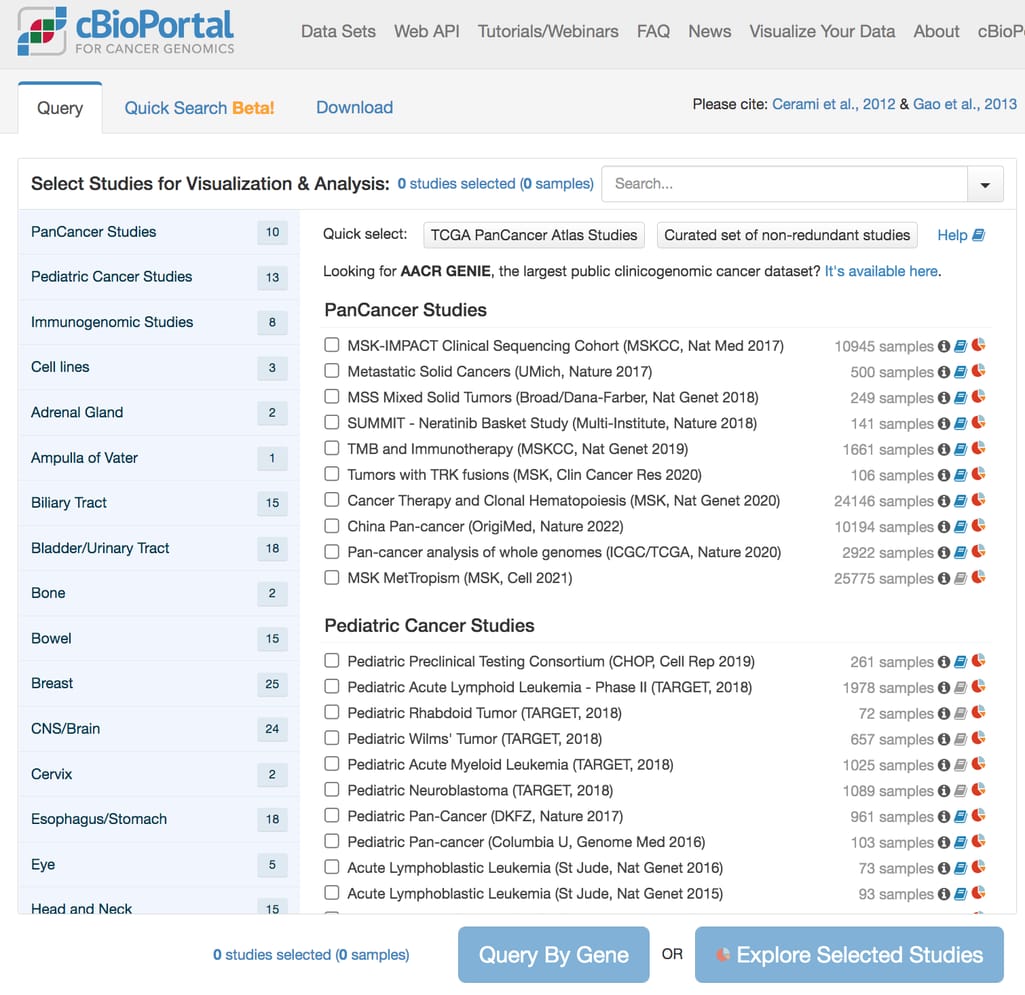 cBioPortal interface
cBioPortal interface
cBioPortal is currently used in over 70 public and private institutions in the world including John Hopkins University, AstraZeneca, MayoClinic, and many more. To use it, download cBioPortal for free on GitHub. They also have an API and R/MATLAB interfaces.
UCSC Xena
The Xena Functional Genomics Explorer by the University of California Santa Cruz is a web tool allowing for exploring and visualizing datasets from UCSC databases such as TCGA, ICGC, TARGET, GTEx, CCLE, and others.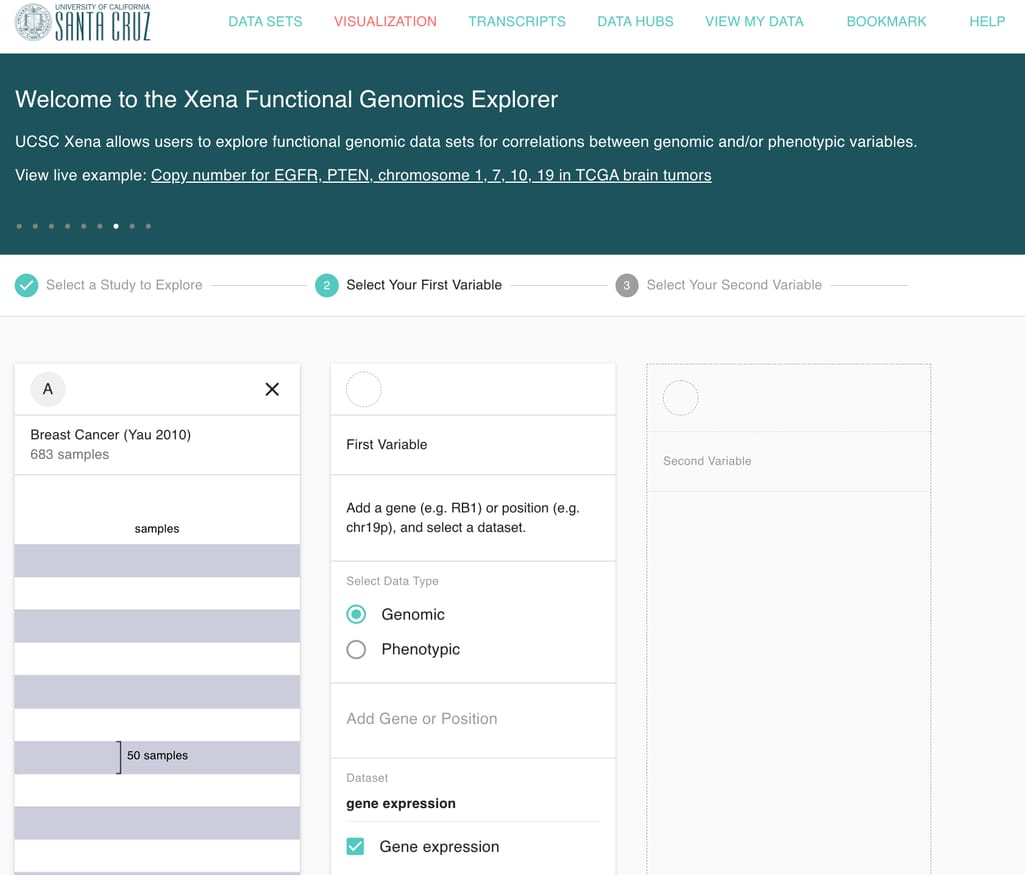
UCSC Xena interface
Xena supports any functional genomics data, including:- single-nucleotide variants (SNVs),
- Insertion–deletion mutations (INDELs),
- large structural variants,
- copy number variation,
- gene-, transcript-, exon-, miRNA-, LncRNA-, protein-expressions,
- DNA methylation,
- ATAC-seq signals,
- phenotypic/clinical annotations, and
- higher-level derived genomic parameters.
LinkedOmics
LinkedOmics is a web application, where you can analyze multi-omics cancer data from 32 cancer types from TCGA and 10 CPTAC cancer cohorts.The portal includes such data types as:
- data from methylation,
- copy number variation, mutation,
- mRNA expression,
- miRNA expression,
- RPPA, and
- clinical data (phenotype) related to primary tumors from 11,158 patients.
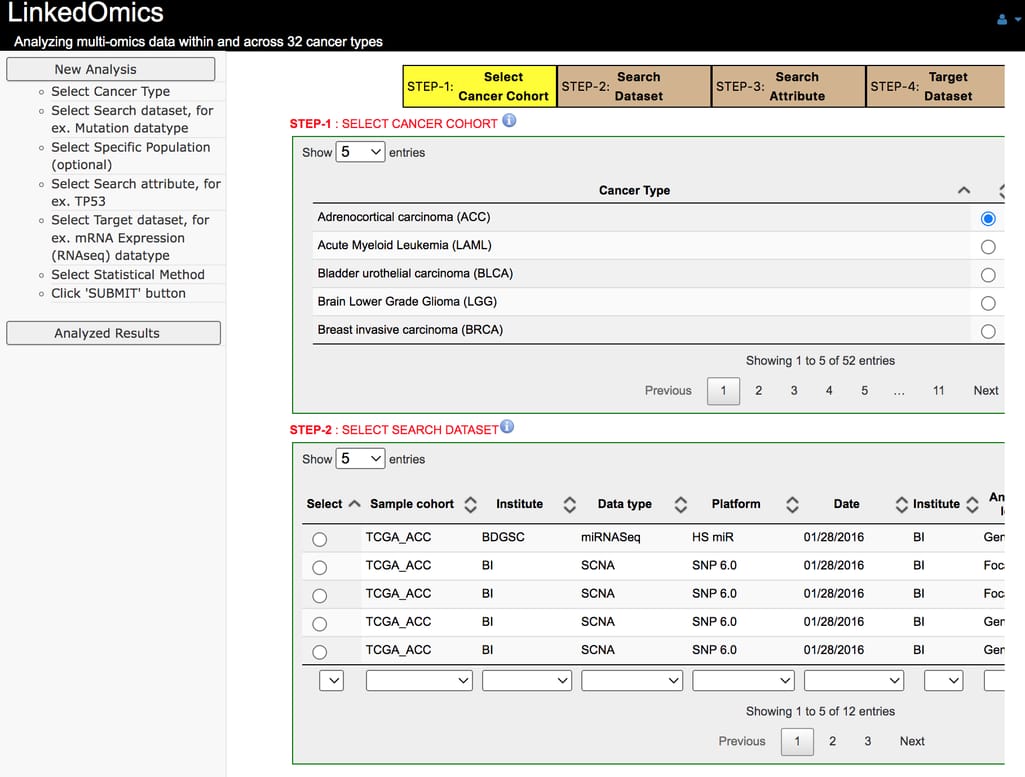
LinkedOmics interface
LinkedOmics has three analytical modules: LinkFinder to search for attributes, LinkInterpreter to perform analysis, and LinkCompare to perform meta-analysis based on results generated by LinkFinder.NetGestalt
NetGestalt is a web app for multi-omics data visualization and integration. What’s special about it is that it visualizes on a one-dimensional layout, allowing simultaneous presentation of large-scale data from various sources. It uses NetSAM, an R package, to perform network seriation and modularization analysis.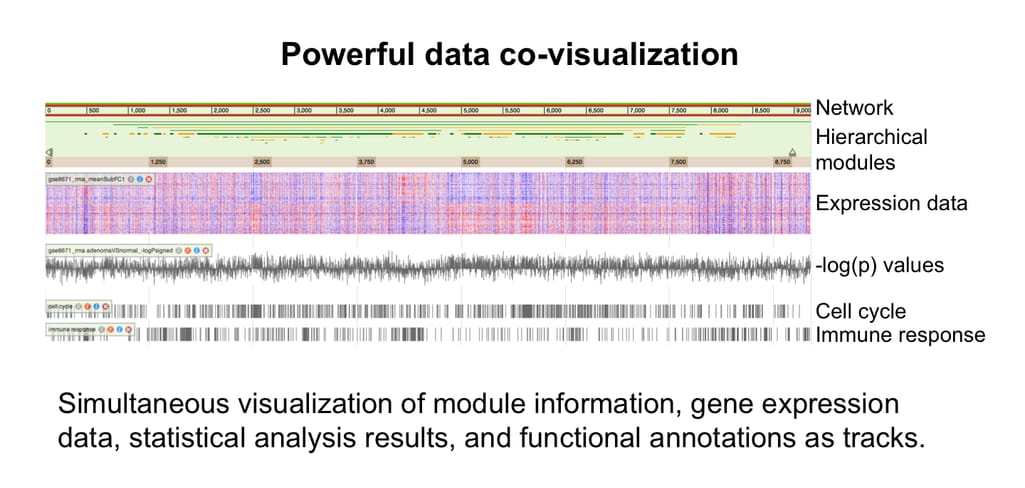
NetGestalt capabilities
It accesses datasets from TCGA, CPTAC, and Colorectal Cancer Portal or you can integrate your own data (both human and mouse).PaintOmics
PaintOmics is a web tool for integrated analysis of multi-omics data using biological pathway maps. The tool was developed to accept data of a diverse nature and supports visualization into KEGG, Reactome, and MapMan pathway maps for more comprehensive knowledge of multiple species of different biological kingdoms.PaintOmics accepts the following input data:
- gene expression data, for example, mRNA-seq or microarrays,
- metabolite data, and
- region-based data, like ChIP-seq and DNase-seq data.
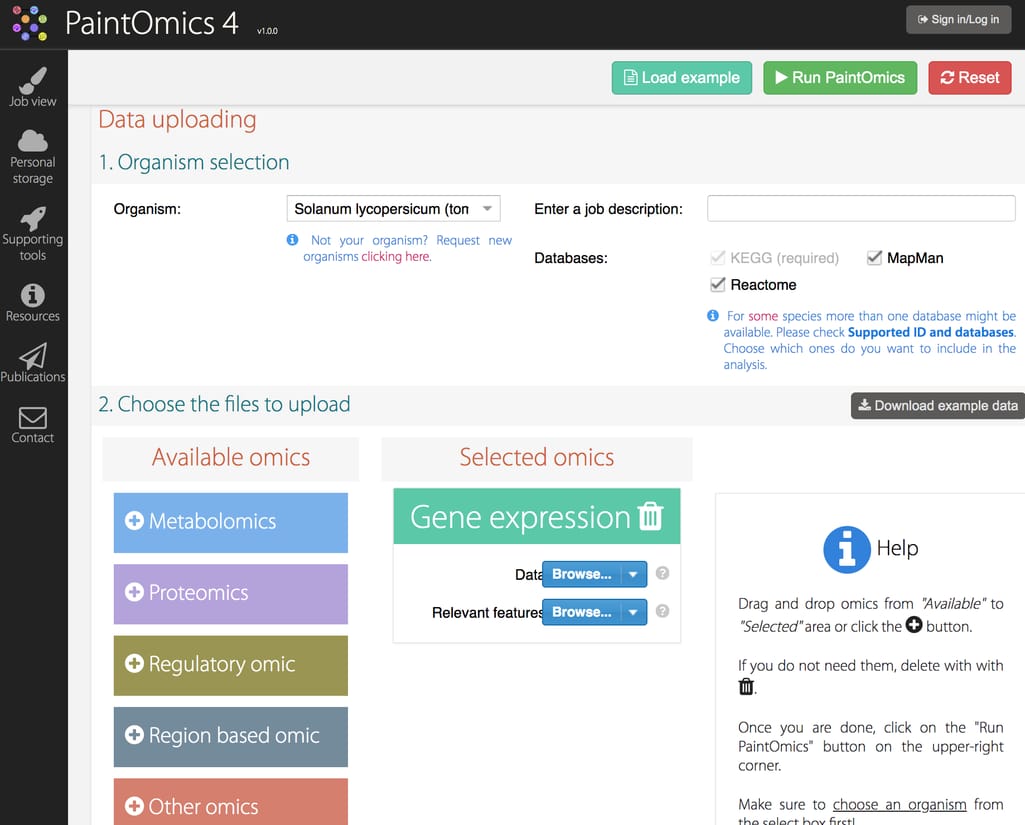
PaintOmics 4 interface
PaintOmics analyzes only user input data. Users should provide two files for each omic data type: a file with measurements and a list of relevant features.Challenges of omics data analysis and how to approach them
The diversity and volumes of omics data make it a subject of big data analytics and require the use of advanced computational methods such as machine learning and deep learning. Unfortunately, just as it’s useful and integral to modern scientific research, the adoption of omics data also presents challenges that don’t currently have standardized solutions.If we talk about multi-omics data, which combines datasets of different omics groups for even a more holistic view of human health, the situation is even more difficult.
So omics data may be challenging to use for the following reasons.
The curse of dimensionality
This phenomenon from data science refers to the problem when the number of variables in the dataset exceeds the number of samples, which creates data sparsity for an ML model. In biological studies, this happens when you gather a substantially large amount of data from a small sample of patients. The problem grows in multi-omics studies when we take measurements from multiple platforms, significantly increasing the variable number.So omics datasets often need to go through dimensionality reduction either by feature extraction (creating new features from the existing ones) or feature selection (identifying only relevant features).
Missing data
Generally, it’s a common problem for omics datasets to have missing values, for instance, when the test that can obtain that data is costly and available only for a fraction of samples. But the problem is highlighted in multi-omics analysis, where there can be even more samples with missing data.To handle this problem, there are various methods you may apply. There’s listwise deletion, meaning excluding the entire sample if the data for any variable is missing. Of course, this leads to substantial information loss. Another method is called imputation, where a regression model replaces missing values with a mean or median of the available variables. Read our article on synthetic data to learn about modern approaches to handling the missing value problem.
Hosting and accessing multi-omics data
There’s currently no unified public infrastructure for hosting multi-omics data. There are various omics data repositories but there are no consistent standards and protocols, making it difficult to find and access it.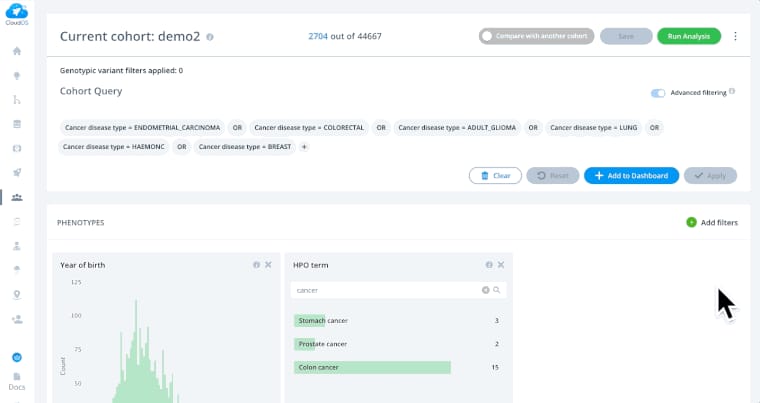
Lifebit CloudOS interface Source: Lifebit
There are commercial products like ones by Lifebit or SevenBridges that provide cloud platforms to host your omics data. Lifebit, for instance, has some open data available for analysis. But there’s still work to do in terms of addressing all possible study designs.Big data scalability
Normally, machine learning models improve their performance with the availability of more data. But when data is acquired via multi-omics platforms, scalability challenges may rise. For instance, it’s unfeasible to analyze multi-omics using ML methods on a single computer. Or sometimes you can’t train models on the entire dataset at once. Also in some studies, training samples would arrive over time, so streaming analytics must be applied.For such large scale analyses, novel computationally efficient algorithms and cloud computing solutions like ML as a service are being applied
Heterogeneous data
Multi-omics analyses commonly suffer from data heterogeneity, when datasets have different numbers of variables and mismatched scaling, and are generated using different formats and platforms. Integrating these diverse data types into a single prediction model is challenging.Decision trees, regression models, and neural networks are all used to combat this issue, as well as networks like HeteroMed, PARADIGM, or SNF that can handle raw text, numeric, and categorical data in electronic health records (EHRs).
Joining the progress
The advent of multi-omics research has resulted in the most comprehensive understanding of human disease we’ve ever experienced. Once biology continues implementing and embracing data science, we will be able to predict disease advancements with unmatched accuracy, discover new, more effective drugs, and prevent suffering for millions of people worldwide.But although you can use many data analysis platforms without programming or data science knowledge, those skills are integral for preprocessing data, interpreting it, and driving the research forward. In just a few years, we won’t be able to separate healthcare from computer science, so learning to operate big data must become your priority soon.

Maryna is a passionate writer with a talent for simplifying complex topics for readers of all backgrounds. With 7 years of experience writing about travel technology, she is well-versed in the field. Outside of her professional writing, she enjoys reading, video games, and fashion.
Want to write an article for our blog? Read our requirements and guidelines to become a contributor.

